How I broke tf.data.datasets down into manageable chunks
Relative Performance of .shard(), .skip() and .batch() in TensorFlow
 Source: Eric Nielson (https://unsplash.com/photos/QdcaEK4B1xw)
Source: Eric Nielson (https://unsplash.com/photos/QdcaEK4B1xw)
Ever have the wonderful experience of a multi-day/week processing job crashing
on you at 99%, only to have to re-process everything again? Read on to find
out how I managed to split a tf.data.dataset into smaller chunks, and the
relative performance of the various approaches I tried.
TL;DR
In general, prefer using .batch() over .shard() or .skip() to split
tf.data.dataset into chunks for processing / saving of progress etc.
Background
As part of my day job at the National Neuroscience Institute, I work with medical images. Properties of such images are two-fold:
-
First, each image is 3D;
-
Secondly, each image may be of very high resolution.
As a result, even with ~400 samples, I would have data in the hundreds of gigabytes.
Initial Googl-ing
There must be a better way to handle the data, I thought to myself, picturing the stereotypical TV informercial actor/actress scratching his/her head.
My initial Gut Feel™ is that the data should be split into smaller files, and a
quick search confirmed my belief as to why it may be a good idea to split the
.tfrecord files:
https://datascience.stackexchange.com/questions/16318/what-is-the-benefit-of-splitting-tfrecord-file-into-shards.
Next, I needed a way to actually split my data files, and a brief Google session
landed me on this solution which suggested the use of .shard().
-
Naturally, I copied and pasted the code into my notebook, selected
Restart kernel and ran all cells, and lived happily ever after. -
Of course that is not the case. My code ended up taking way longer to run, and I had to kill the job.
Having no other obvious choiceI decided to do some investigation, and below are the totally non-vigorous and unscientific results:
Performance Testing Pre-requisite
I first coded up an timer to keep track of the time:
@contextlib.contextmanager
def timed():
import datetime as dt
start = dt.datetime.now()
try:
yield None
except:
raise
finally:
end = dt.datetime.now()
print(f'Duration: {end - start}')-
Some Notes on the above fragment:
-
The
@contextlib.contextmanagerautomagically converts a generator function into a resource manager, perfect for a use-case like ours. I would strongly recommend reading up on it and keeping it under your toolbelt. -
The
import datetime at dtis put within the function so that the function may be part of a bigger utilities module, without needing the module to import everything at initialization. Also, since Python caches it's import, thedatetimemodule will only be imported once in any case.
-
The timer function may be used as so:
with timed():
runAllTheMethods()Performance Baseline
Next, we need a baseline measure.
-
This baseline measure will be compared against the performance of each of the various approaches to splitting the dataset.
-
The comparison will allow us to evaluate whether the act of splitting the dataset itself results in slower processing of the data.
The baseline is obtained by iterating through the whole tf.data.Dataset
without any splitting or doing anything else:
test_ds = loadMyDataset()
NUM_TRIALS = 5
for i in range(NUM_TRIALS):
with timed():
for _ in test_ds:
pass # In real application, actual processing is done here
.skip()
First, we test the performance of .skip().
-
We do so by iterating through the
tf.data.Datasetafter using.skip()to skip all but one element:for i in range(NUM_TRIALS): with timed(): for _ in test_ds.skip(NUM_DATA-1): pass # In real application, actual processing is done here -
The hypothesis is that since we are skipping all elements but one, it should be much faster than the baseline of iterating through every record.
-
If the hypothesis is true, then we will proceed to use
.skip()together with.take()to split the data.-
For example, assuming each split has
10records,ds.skip(0).take(10)will give the first split, andds.skip(10).take(10)will give the second split, and so on. -
The example code would look like this:
NUM_TRIALS = 5 NUM_SHARDS = 5 items_per_shard = math.ceil(NUM_DATA / NUM_SHARDS) for _ in range(NUM_TRIALS): with timed(): for i in range(NUM_SHARDS): for _ in test_ds.skip(items_per_shard*i).take(items_per_shard): pass
-
.shard()
Next, we test the performance of .shard().
-
We do so by iterating through the
tf.data.Datasetafter sharding into five shards:NUM_TRIALS = 5 NUM_SHARDS = 5 for _ in range(NUM_TRIALS): with timed(): for i in range(NUM_SHARDS): for _ in test_ds.shard(NUM_SHARDS, i): pass # In real application, actual processing is done here -
Since we are still going through the whole
tf.data.Datasetonce, the time taken should be comparable to the baseline, since both would iterate through every record.
.batch()
Finally, we test the performance of .batch().
-
We do so by iterating through the
tf.data.Datasetby manually splitting into five batches:NUM_TRIALS = 5 NUM_SHARDS = 5 items_per_shard = math.ceil(NUM_DATA / NUM_SHARDS) for _ in range(NUM_TRIALS): with timed(): for shard in test_ds.batch(items_per_shard): for _ in shard: pass # In real application, actual processing is done hereNote in this approach that we do need to know the total number of data (i.e.,
NUM_DATA) in order to split into a specific number of files. -
Since we are still going through the whole
tf.data.Datasetonce, the time taken should be comparable to the baseline, since both would iterate through every record.
Results
The results are as follows:
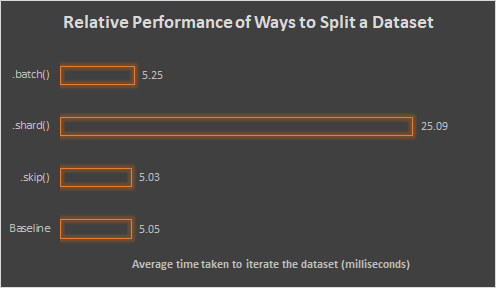
Analysis:
-
Comparing
.skip()and the baseline, we can see that even though we skipped all the records but the last one, it still took essentially the same amount of time as compared to the baseline which iterate though each records.-
This suggests that skipping a record still incur the costs of the earlier processing steps in pipeline. I.e., TensorFlow is not doing anything clever to avoid the skipped item right from the start.
-
-
Comparing
.shard()and the baseline, we can see that.shard()is taking 5 times as long as the baseline.-
Recall that we are spliting the data into 5 shards, which correspond to the 5 times duration taken by
.shard(). -
This suggests that for each shard, the
.shard()operation is nonetheless processing the whole pipeline. -
This makes sense because TensorFlow's documentation on tf.data.Dataset.shard() states that sharding is done by splitting records into groups by taking the modulo of each record's index—to get each record index, it must naturally iterate through the whole dataset.
-
-
Finally, comparing
.batch()and the baseline, we can see that both takes about the same amount of time.-
This suggests that using
.batch()to split the dataset will incur little to none overhead.
-
!!! Conclusion: Generally prefer .batch() when splitting your dataset to avoid
unnecessary overheads.
Sidenote:
-
In the above test conducted, the
test_dsrepresents a pipeline that involves processing steps like loading images from file, scaling and normalization.-
These processing steps are likely what is being duplicated in the
.skip()and.shard()cases, resulting in poorer performance.
-
-
If the
.skip()and.shard()method were applied at an earlier stage—say, immediately after globbing a list of file names—then the performance difference will be much less pronounced.-
That said, it would generally be difficult to plan in advance at an early stage of the processing pipeline how we want to split the data. And in some cases where we get the dataset from another sources, it might not be possible to easily insert the
.skip()or.shard()early in the pipeline.
-
Lessons Learnt
Don't just copy and paste code from StackOverflow. Easy to say; hard to follow.
Use .batch() to split your datasets. Or use .skip() / .shard() at earlier
on in the pipeline, before any heavy processing.